The location-scale regression model assumes a normally distributed response variable with one linear predictor for the mean (= the location) and one for the standard deviation (= the scale). The standard deviation is mapped to the linear predictor through a log link.
This function sets up the model object and estimates it with maximum likelihood.
Usage
lmls(
location,
scale = ~1,
data = environment(location),
light = TRUE,
maxit = 100,
reltol = sqrt(.Machine$double.eps)
)Arguments
- location
A two-sided formula with the response variable on the LHS and the predictor for the mean on the RHS.
- scale
A one-sided formula with the predictor for the standard deviation on the RHS.
- data
A data frame (or list or environment) in which to evaluate the
locationandscaleformulas.- light
If
TRUE, the design matrices are removed from the estimated model to save some memory.- maxit
The maximum number of iterations of the Fisher scoring algorithm.
- reltol
The relative convergence tolerance of the Fisher scoring algorithm.
Value
A fitted linear model for location and scale as an lmls S3 object.
The object has at least the following entries:
y: the response vectornobs: the number of observationsdf: the degrees of freedomdf.residual: the residual degrees of freedomcoefficients: the regression coefficients as a list with the nameslocationandscalefitted.values: the fitted values as a list with the nameslocationandscaleresiduals: the response residualscoefficients: the variance-covariance matrices of the regression coefficients as a list with the nameslocationandscaleiterations: the number of iterations the Fisher scoring algorithm took to converge
Examples
library(lmls)
m <- lmls(y ~ poly(x, 2), ~ x, data = abdom)
summary(m)
#>
#> Call:
#> lmls(location = y ~ poly(x, 2), scale = ~x, data = abdom)
#>
#> Deviance residuals:
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> -3.363000 -0.701400 -0.048870 -0.000149 0.623200 4.066000
#>
#> Location coefficients (identity link):
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 226.734 0.563 402.739 < 2e-16 ***
#> poly(x, 2)1 2160.371 15.228 141.871 < 2e-16 ***
#> poly(x, 2)2 -99.184 12.447 -7.968 1.61e-15 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Scale coefficients (log link):
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 1.356462 0.096714 14.03 <2e-16 ***
#> x 0.042291 0.003388 12.48 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual degrees of freedom: 605
#> Log-likelihood: -2396.41
#> AIC: 4802.82
#> BIC: 4824.89
#>
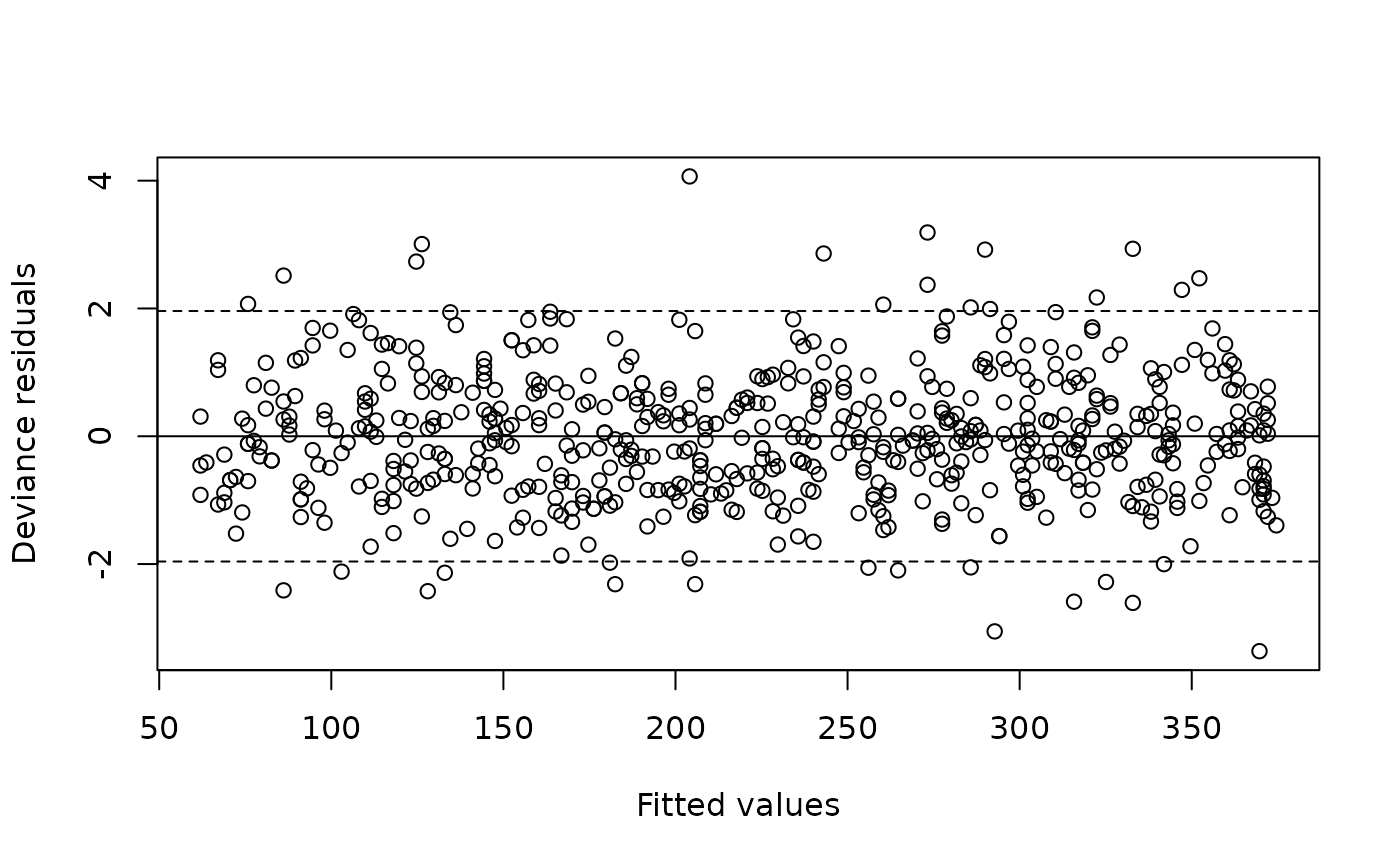
plot(m)
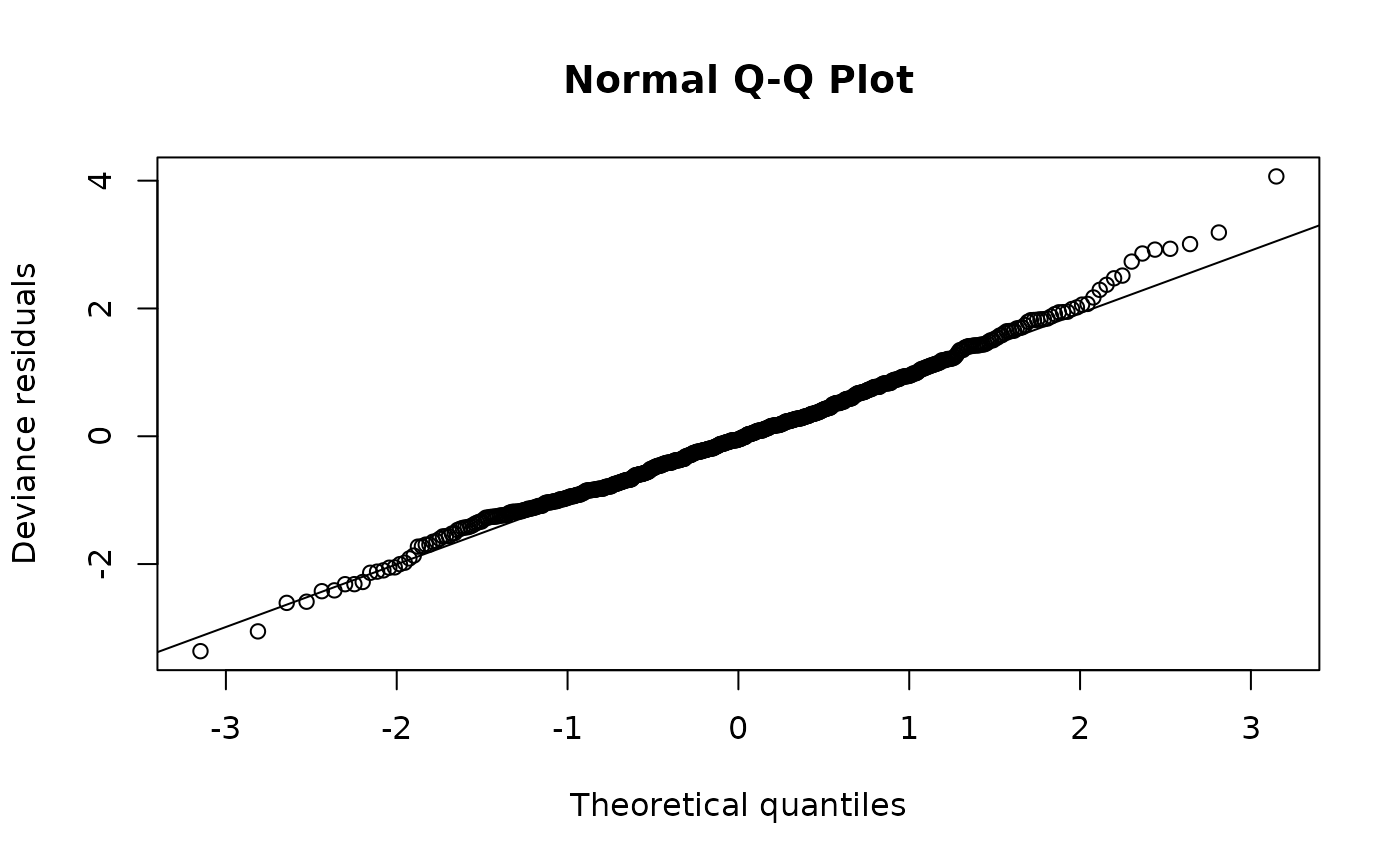
 qqnorm(m)
qqnorm(m)